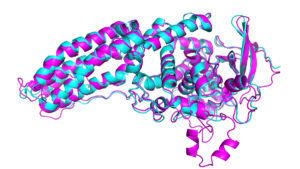
Proteins are long, convoluted molecular chains called macromolecules. They are enzymes, that catalyse almost every chemical reaction needed to keep the body alive. E.g. thyroxine plays vital role in digestion, heart and muscle function, haemoglobin carries the oxygen, immunoglobulin is responsible for defence in the body and keratin for hair and skin (SARS-CoV-2 spike a protein, that allows coronavirus to invade human cells). Each one of them is built from a menu of 21 different types of amino acids. But they are not the main problem. Machines which were able to analyse composition of amino-acid have existed for several years. But the functioning of the protein is also defined by its shape. For now we can only try to shape them using X-ray, but it is a very time-consuming process. Today, on November 30th biologists from DeepMind – laboratory owned by Google showed howto use a computer to predict those shapes. The measure now used in checking the progress is Critical Assessment of Protein Structure Prediction (CASP). “The algorithms are subjected to blind test their abilities to predict shapes of several proteins of known structure”. The first try DeepMind did was 2 years ago and the progress they have achieved is extraordinary. AlphaFold 2 compares the presumable locations of the atoms to the real ones and it has an average score of 92.4%. 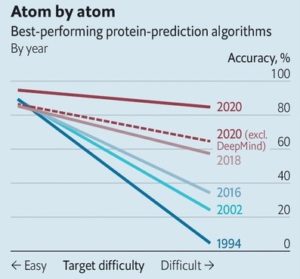
Why did DeepMid performed so well at this? The answer is in what they were doing before this project. Their best known achievement is for AlphaGo. Their programmed a computer (“taught”) how to play a game named Go. Go is a game created in China over 2,500 years ago. It is believed to be the oldest board game in the world and the aim is to surround more territory, than enemy. Lee Sedol – one of the world’s best players has been defeated by AlphaGo in 2016. The game has around 10^170 positions, whereas it is believed to be 10^300 different shapes in which a protein could find itself. Because of such a large number the inventors had to go a way they took in Go project, which is looking for shortcuts. Of course players are not machines. They depend on their intuition, general understanding of the game and their own strategy. It may seems tough to teach a machine how to play, but the answer is examples. AI is know from self-learning abilities. When we give a machine enough examples of folding proteins, it will learn the shortcuts and some rules humans do not even notice they apply. This scientific research can boost drugs invention and can also help with numeral diseases, which are caused mainly by misshapen proteins, such as Alzheimer’s, Parkinson’s, Creutzfeldt–Jakob’s disease, Huntington’s and many others.
Sources:
https://deepmind.com/blog/article/AlphaFold-Using-AI-for-scientific-discovery
https://www.theguardian.com/technology/2020/nov/30/deepmind-ai-cracks-50-year-old-problem-of-biology-research
https://www.nature.com/articles/d41586-020-03348-4
https://www.sciencedirect.com/topics/neuroscience/protein#:~:text=Proteins%20are%20macromolecules%20formed%20by,chains%20to%20form%20a%20protein.